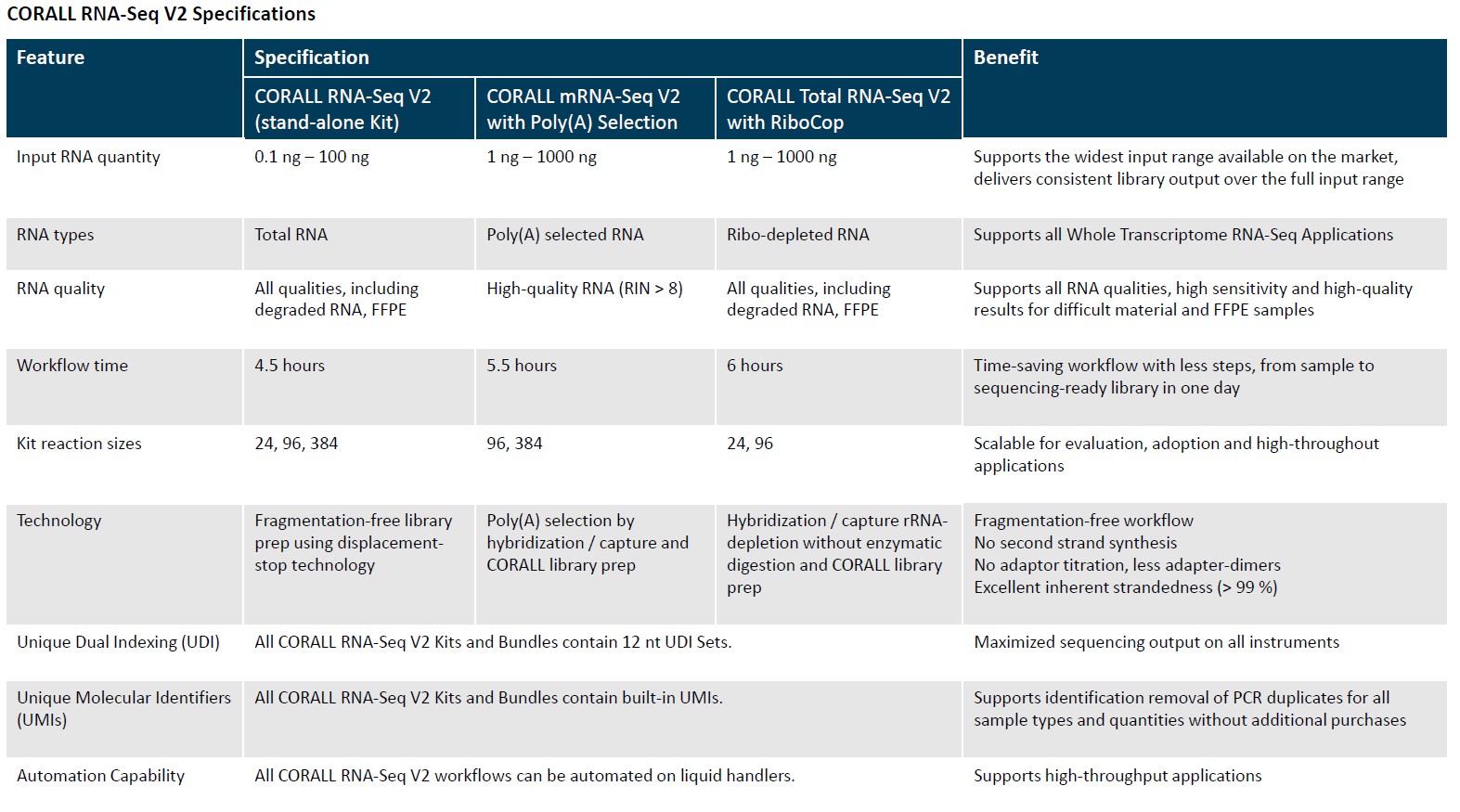
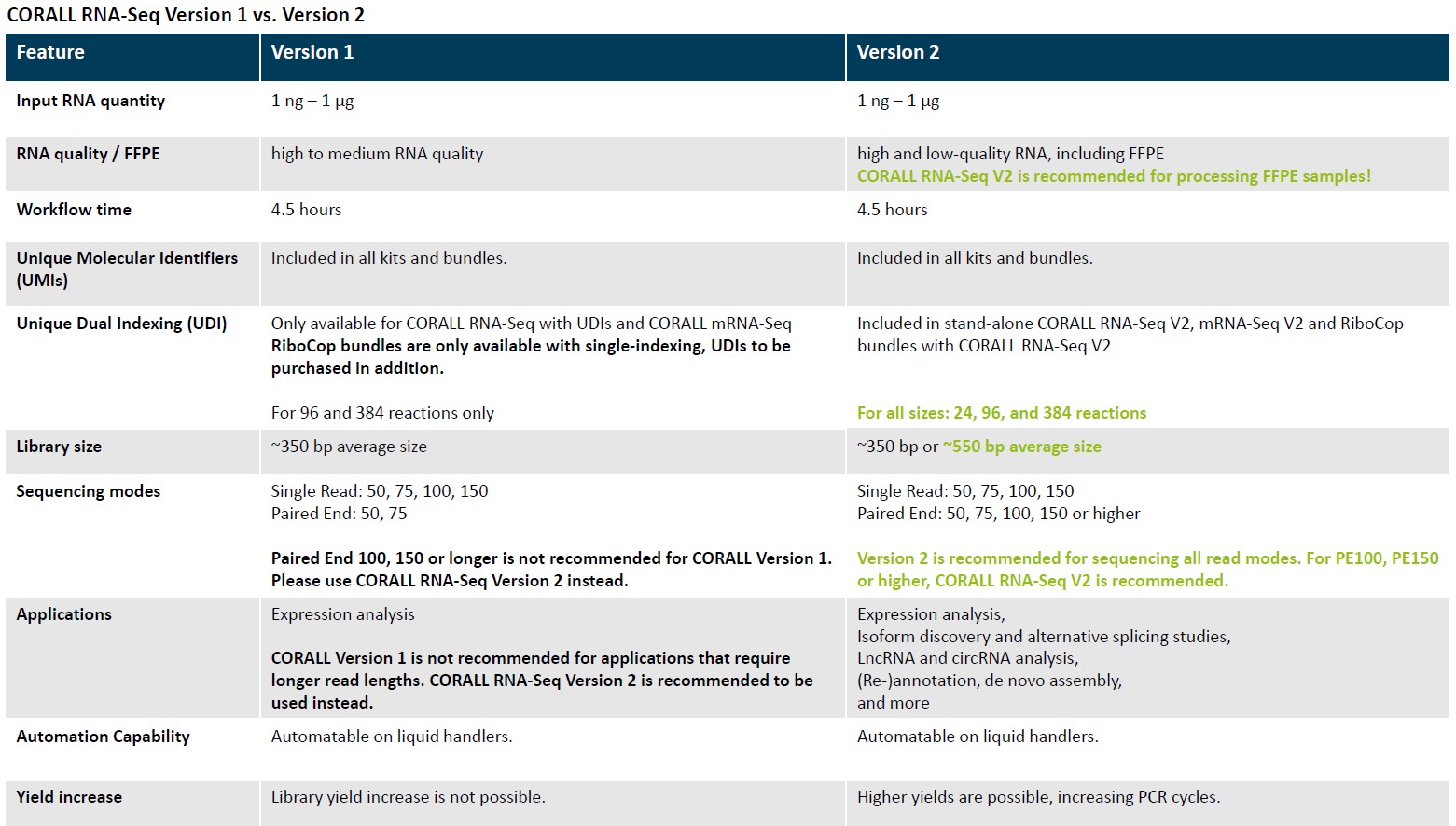
The CORALL RNA-Seq Library Prep Kits enables fast and cost-efficient generation of stranded, UMI labelled, and unique dual indexed libraries for whole transcriptome analyses using Illumina® NGS platforms.
The Lexogen Total RNA Library Prep kits have been well established over the last years and have proven to deliver high quality RNA-seq libraries, generating best-in-class results.
In addition, CORALL RNA-seq Library Prep kits succeed in consolidating your RNA-seq workflows, as the CORALL kits can be used for mRNA-seq as well as for total RNA-seq, simply by adding a specific module to treat your input material.
Product information
Check now the NEW VERSION!!