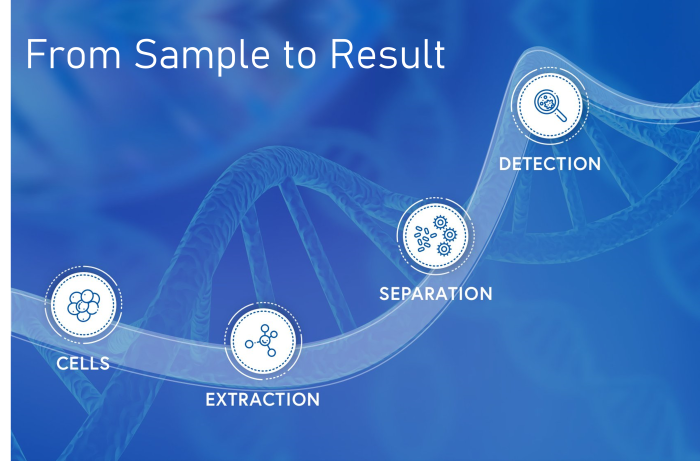
- NanoDrop micro-volume spectrophotometers
- EzDrop Micro-volume spectrophotometers
- Microplate readers
- NextGenPCR Machine
- PCR Thermal Cyclers: TurboCycler
- 3D Cell Culture: Functional Organoids Growth
- Automated DNA/RNA Extraction system
- Automated systems
- Automated Western Blot Processor
- Bioreactors
- Capillary Electrophoresis Analysers
- Cell Counters
- Electrophoretic Automated Tissue Clearing
- Imaging Systems
- Next Generation icIEF
- UV-Vis Spectrophotometers and Fluorometer
- Small laboratory equipment