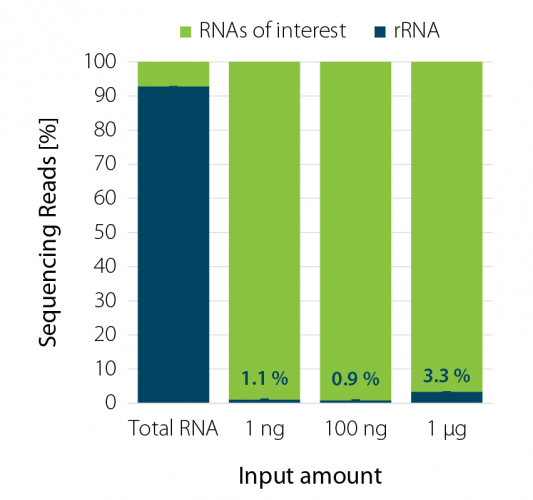
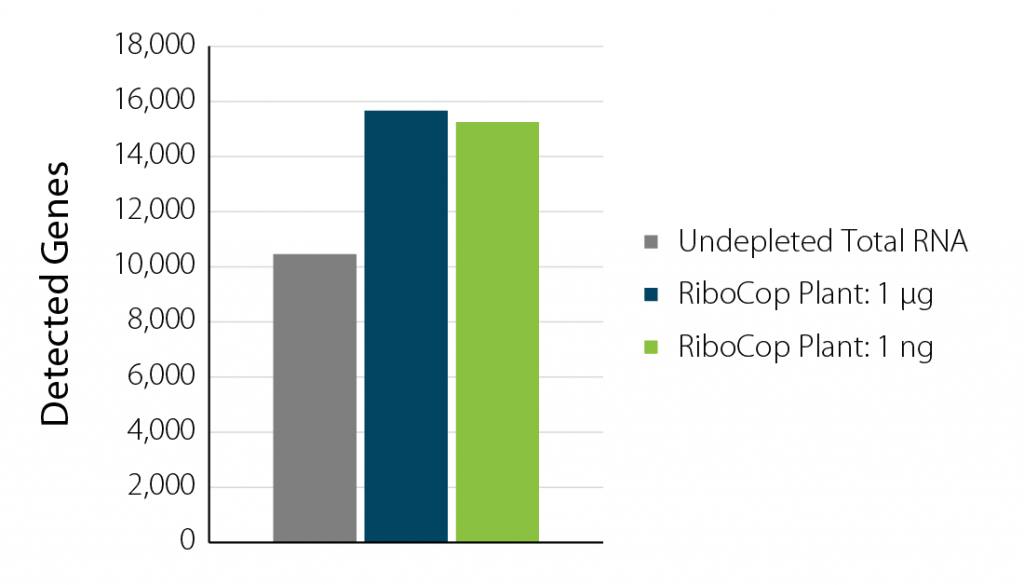
When looking at total RNA, about 80% is ribosomal RNA (rRNA) and is mostly unwanted in RNA-seq experiments. When rRNA is not removed from the sample during library prep, it will occupy most of the sequencing space, reducing the number of samples that can be pooled on a single lane of a flow cell and therefore, increase the sequencing cost dramatically.
Next to total RNA-seq, mRNA-seq is also an important application. Knowing that the mRNA-fraction only represents a few percentages of the total RNA, it is good practice to enrich for this fraction prior to mRNA library prep by selecting and capturing the poly-A tailed mRNA.
We do offer 2 different options for rRNA depletione or Poly-A selection. Combined with the CORALL total RNA-seq library prep kit, this represents a powerful and streamlined method for total RNA- and mRNA-seq